Description:
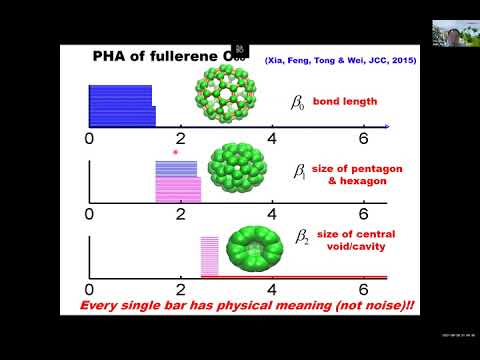
Explore a 47-minute conference talk on persistent function-based machine learning for drug design. Dive into the development of new persistent functions like persistent spectral, persistent Ricci curvature, and hypergraph-based persistent homology. Learn how these representations generate molecular descriptors and fingerprints, combining with machine learning models such as random forest, gradient boosting tree, and convolutional neural networks. Discover how these molecular features characterize intrinsic multiscale topological information within molecules, offering better transferability for machine learning models. Examine the performance of these models tested on PDBbind-2007, PDBbind-2013, and PDBbind-2016 databases, outperforming traditional molecular descriptor-based machine learning models. Gain insights into biomolecular data, learning models, energy curves, molecular dynamic simulations, weighted homology models, and topology-based models. Investigate spectral models, prediction comparisons, global learning, hybrid models, and the application of mathematical AI in molecular science.
Read more
Kelin Xia - Persistent Function Based Machine Learning for Drug Design
Add to list
#Science
#Chemistry
#Drug Design
#Computer Science
#Machine Learning
#Mathematics
#Geometry
#Topology